Our long-term collaboration with Insilico Medicine, a company that focuses on artificial intelligence for drug discovery and longevity research, has borne some very important fruit. We have released a benchmarking platform for generative models, which we named MOSES (MOlecular SEtS). You can find the paper currently released on arXiv, the github repository, and the press release by research partner Insilico. Congratulations to the whole team! Before we dive into a little bit of detail, here is the Neuromation staff together with our collaborators from Insilico at the NIPS (NeurIPS, as they call it now) conference currently held in Montreal:

So what is MOSES, why do we care, and what do Neuromation and Insilico have in common here?
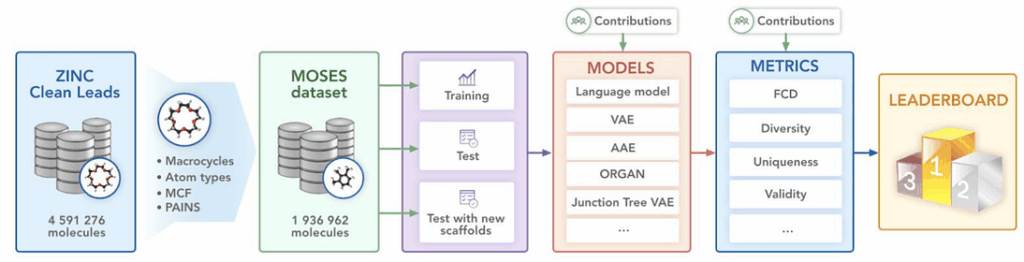
MOSES is a benchmarking platform for generative models that aim to generate molecular structures. We covered generative models such as generative adversarial networks (GANs) before (e.g., here or here). In molecular biology and biochemistry, generative models are used to produce candidate compounds that might have desired qualities. We have already published a post about our previous joint project with Insilico which provides more details about such models.
One common thread in generative models is that they are really difficult to evaluate and compare. You have a black box that produces, say, images of human faces. Scratch that, you have twenty black boxes. Which one is best? You could try to get closer to the true answer by asking real people to evaluate the faces, but this surely won’t scale.
So researchers have been developing metrics to compare generative models. I will save a more detailed explanation of the metrics for an in-depth Neuromation Research post which is going to follow soon. For now, let me just say that there are plenty of different metrics. It’s really hard to collect them all from very different implementations, and even harder to claim that your numbers are really comparable with the numbers in other papers. The whole field could sure use some standardization and streamlining.
That’s where MOSES comes in. In this project, we:
- prepared a large dataset of approximately 2 million molecules based on specially designed chemical filters;
- implemented the most popular metrics for the evaluation of generative models;
- most importantly, implemented several state of the art models and provided a large and unified experimental comparison between them.
Building MOSES was a big project. We have been working on it for the better part of this year; 40 weeks in the title are no exaggeration. In a project like that, you need a deep and well integrated collaboration between chemists, medical researchers, and machine learning gurus. And that is exactly what we had between Neuromation, Insilico Medicine, the Harvard University and the University of Toronto.
The result is a benchmarking dataset, an evaluation pipeline, and a large-scale experimental comparison that provides the stable footing so badly needed for this field. Now, new works can build upon this foundation, compare new models with baselines from our paper, and make direct quantitative comparisons in terms of various evaluation metrics. We hope that researchers all over the world will benefit from our joint effort.
My congratulations to our team and to our dear friends at Insilico Medicine! Lots of contributors, but let me please highlight and thank the following individuals: big thanks to Daniil Polykovsky, Alexander Zhebrak, Vladimir Aladinskiy, Mark Veselov, Artur Kadurin, and Alex Zhavoronkov from Insilico, to Benjamin Sanchez-Lengeling from Harvard, Alan Aspuru-Gusik from the University of Toronto/Vector Institute, and to Neuromation researchers Sergey Golovanov, Oktai Tatanov, Stanislav Belyaev, Rauf Kurbanov, and Aleksey Artamonov. Thanks guys!
Thank you for reading and stay tuned for the next updates from our Neuromation Research blog!
Sergey Nikolenko
Chief Research Officer, Neuromation
Leave a Reply